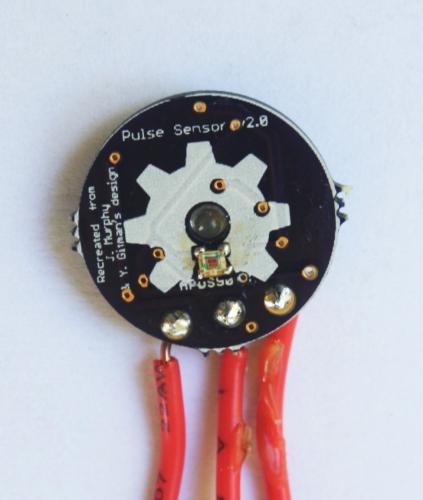
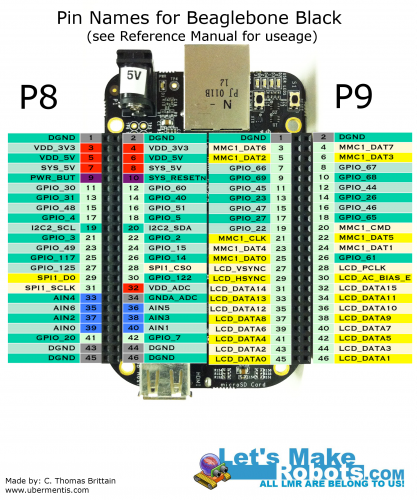
Originally posted on www.letsmakerobots.com
I've been working on this one in silence for a bit.
Awhile back it hit me, before I started growing my Overlord project in complexity I wanted to refine it for ease-of-use. Therefore, I began translating my Overlord project into a Python module I could build off.
I figure, this would make it easier for anyone to use. This includes myself, I've not forgotten my identity as a hack, nor will anyone who pops the hood on this module :)
But, at its core, there are few essential inputs:
- Color to track ...